Let us introduce you to the first production version of Plant.id Health Assessment.
During the last year, we have been collecting feedback on the beta version of our plant disease identification API. Moreover, we have expanded our datasets and deepened our understanding of machine learning-powered plant disease identification.
Here is a list of changes coming with the Health Assessment production release:
- Better accuracy and coverage error
- Improved list of disease classes
- Discarding parent classes
- Separate endpoint for Plant.id Health Assessment
- Localization

Better accuracy and coverage error
If you can’t measure it, it doesn’t exist!
We made a few changes in the model architecture and training data. These changes included:
- improved hierarchy and the list of classes
- new training data (new images of sick plants carefully annotated by our plant pathologists)
- new backbone model and improved neural network architecture.
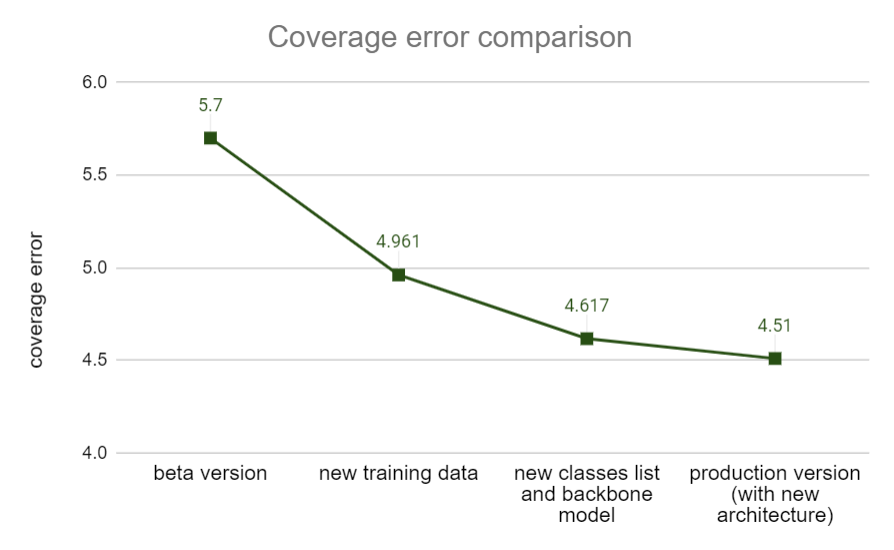
We are pleased to announce that the changes that we made have resulted in increased accuracy. As a result, the top-3 accuracy (the probability of the disease appearing in the top 3 positions in the results) increased from 55.6% to 61.2%. Further, the coverage error which represents an average rank of the correct answer in the response decreased from 5.7 to 4.51.
Improved list of disease classes
Sometimes less is more!
The new model contains 90 disease classes. We had carefully chosen these to resemble the plant health issues of the end-users. We added diseases that were frequent in the health assessment requests we received. For example, mechanical damage and mites. Further, we added diseases that were requested in the feedback such as fungus gnats and springtails. On the other hand, we removed diseases previously underrepresented and thus less important. In some cases, we decided to merge classes with similar concepts and treatments.
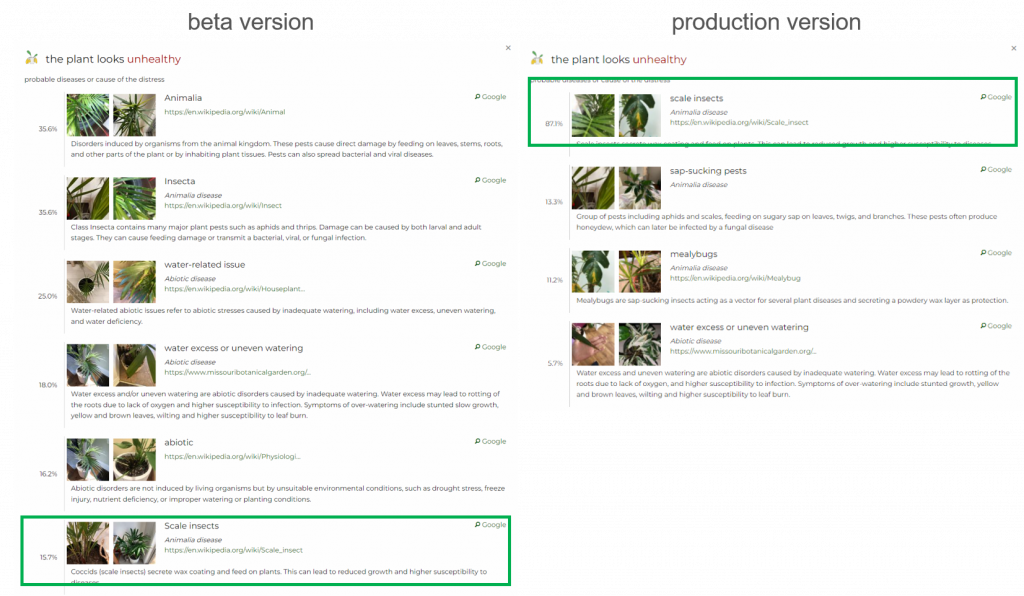
probability (15.7%). Whereas, in the production version, it appears in the first position with a high probability (87.1%).Discarding parent classes
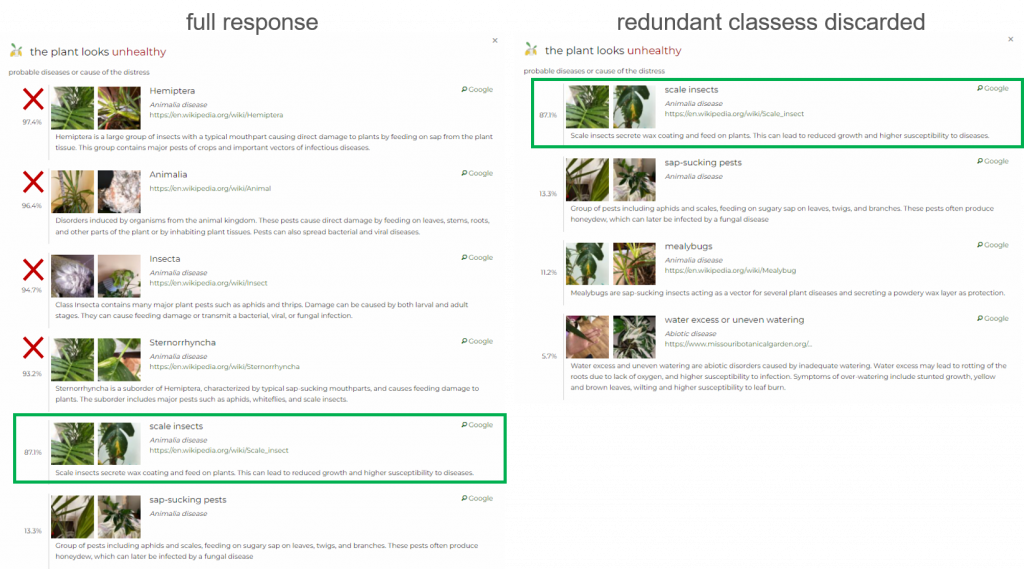
The hierarchical structure of diseases sometimes causes multiple parent-child pairs to appear in the response, providing redundant information.
Now, we offer the possibility to easily discard parent classes. This might be convenient if the difference between probability of the parent class (higher in the hierarchy) and the child class (lower in the hierarchy) is insignificant. In that case, classes that can be discarded gain a boolean attribute redundant in the response.
Specifically, this option becomes available if the probability of the child class is at least 80% of the probability of the parent class. For example, if the probability of a parent class (e.g. abiotic) is 95% and the probability of a child class (e.g. water-related issue) is at least 76%, then the attribute redundant:true will appear in the response.
Try this functionality on our Plant.id Demo page (sign-in required) and explore other improvements that came with the Health Assessment production release.
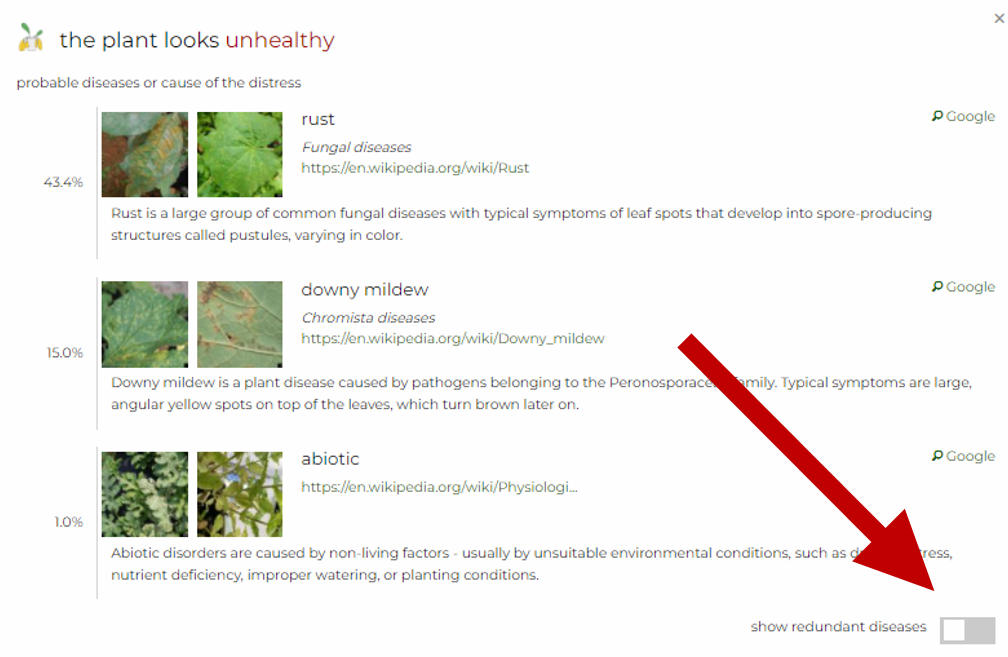
show redundant diseases below the list of diseases.Separate endpoint for Plant.id Health Assessment
We have designed a separate /health_assessment endpoint. This opens up the possibility to use Plant.id Health Assessment independently of species identification.
See the documentation.
Look at the response example.
The pricing is one identification credit per Health Assessment and one per species identification. The base price is €0.05. If you want to know more details about bulk prices, contact us at business@plant.id.
Localization
Gray mold, Grauschimmelfäule or plíseň šedá?
We are going local with Plant.id Health Assessment. The first localization we are preparing is German, but we plan to add more languages in the future.
Localization will be enabled by including the parameter language in the new /health_assessment endpoint. The response will contain information in the selected language, if available. By default, the results will be given in English.
Please, let us know if there is a language you would appreciate on support@plant.id.
