FAQ for Plant.id API
Most frequent questions and answers
For general questions, see the Kindwise web.
Plant.id Species Identification
- How is the identification result constructed?
- What kind of plants can you identify?
- What is the content coverage in language-independent plant details?
- What is the content coverage in a specific language?
- Can you identify plant varieties?
- Can I only get disease identification when a plant is sick?
- What is the total cost of identifying diseases?
- Sometimes the result falsely states that the plant is healthy or detects a wrong disease. Am I doing something wrong?
- Can I get a list of all the diseases that the Plant.id Health Assessment API can identify?
- The suggestions are too general (e.g. abiotic, animalia). How can I improve it?
- What is the difference between crop.health and Plant.id Health Assessment?
Plant.id Species Identification
How is the identification result constructed?
The plant identification process currently involves three independent models:
-
- Genus model (such as Philodendron)
-
- Species model (such as Philodendron hederaceum)
-
- Infraspecies model (such as Philodendron hederaceum var. oxycardium ‘Brasil’)
You can choose which model’s results are shown in the response by using the classification_level parameter.Results from each model are post-processed in several steps where classes may be merged and probabilities adjusted. It involves re-evaluating the response to avoid duplicate information. If you want to omit the postprocessing and get the original response instead, you can set classification_raw=true. See the example.
What kind of plants can you identify?
We can currently identify around 30,000 plant species of various life forms. You can explore how our model performs on houseplants, garden ornamentals, trees and shrubs, weeds, European or American wild plants.
What is the content coverage in language-independent plant details?
See available plant details in the API response in the documentation.
The following table shows the content coverage for classes in the Plant.id database.
| content | coverage |
| url | 100% |
| Taxonomy – Kingdom | 100% |
| Taxonomy – Phylum | 100% |
| Taxonomy – Class | 100% |
| Taxonomy – Order | 100% |
| Taxonomy – Family | 100% |
| Taxonomy – Genus | 100% |
| GBIF id | 99.99% |
| iNaturalist_id | 98.65% |
| Wikipedia image | 99.12% |
| Propagation methods | 9.16% |
| Edible parts | 8.80% |
| Watering | 9.50% |
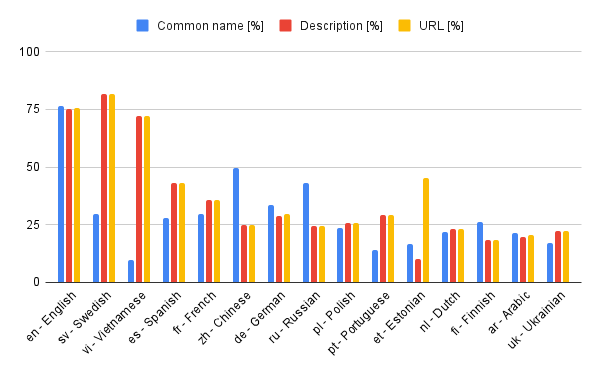
What is the content coverage in a specific language?
Some information (description, common name and URL) is language-dependent. You can set the preferred language in your request (English is the default). The chart below shows the percentage coverage of the top 15 languages (see this sheet for all languages).
Can you identify plant varieties?
Yes, we can! Our latest model update from October 2023 (read a blog post for more) includes a capability to identify so-called infraspecific taxa, which include plant varieties, cultivars and subspecies. However, this is not a default setting of the API; in the Identification request, you need to set the classification_level = all. See the example.
Plant.id Health Assessment
Can I only get disease identification when a plant is sick?
Yes, we have an extra model is_healthy that can identify whether a plant is healthy or diseased. You can use the health = auto attribute to decide whether an image should undergo a Health Assessment automatically. If the model’s output exceeds a specific threshold, indicating the presence of a diseased plant, the image will be sent to the Health Assessment. If the plant is considered healthy, the cost is one credit. If the plant is considered diseased and a Health Assessment is requested, the cost is two credits.
If you want responses from both Species Identification and Health Assessment, you can set the parameter health = all. The total cost would be two credits.
What is the total cost of identifying diseases?
The cost is one identification credit, the same as for plant identification. In case you want both the Species Identification and Health Assessment results for a single plant, the cost is one credit for each product. The base price is €0.05 per credit. Discounts are available for bulk orders. Contact business@plant.id for details.
Sometimes the result incorrectly states that the plant is healthy or identifies the wrong disease. What am I doing wrong?
Diagnosing plant health is often a complex task. For example, if a plant is consistently overwatered, it is also unable to use nutrients effectively, so it will develop symptoms of both overwatering and nutrient deficiency. Therefore, when implementing this functionality, we recommend that you list more than one possible cause of disease in the result.
However, you can also improve the result by taking a photo of a diseased part of a plant for best results. Read our blog post for tips on using Plant.id Health Assessment. Some diseases have less visible symptoms (e.g. small pests), and the detail is crucial for correct identification.
Can I get a list of all the diseases that the Plant.id Health Assessment API can identify?
With Plant disease identification API you can recognize:
-
- 28 classes of fungal diseases
-
- 18 classes of abiotic disorders
-
- 17 classes of pests
-
- 7 classes of chromista
-
- 5 classes of viruses
-
- 5 classes of bacterial diseases
The full list of diseases changes dynamically. If there is a reason you need the full list, please let us know at info@plant.id.
The suggestions are too general (e.g. water-related issue). How can I change this?
You can use the redundant attribute to discard parent classes.
Discarding parent classes is useful when the difference between the probability of the parent class (higher in the hierarchy) and the child class (lower in the hierarchy) is insignificant. In this case, classes that can be discarded have a boolean attribute redundant in the response.
Specifically, this feature in the response activates when the probability of the child class is at least 80% of the probability of the parent class (or less than two percentages in absolute numbers that aim to assess cases with a probability of less than 10%). For example, when the probability of a parent class (e.g. water-related issue) is 95% and the probability of a child class (e.g. water deficiency) is at least 76%, then the attribute redundant=true will appear in the response.
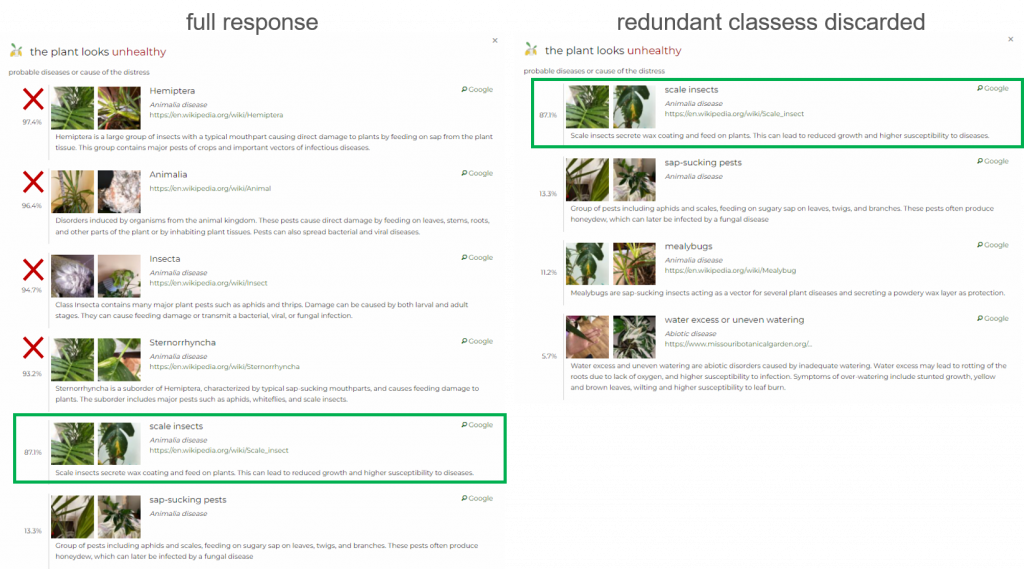
Another option is to use the prune_diseases attribute (default for API v3). This will remove the general classes from the response.
What is the difference between crop.health and Plant.id Health Assessment?
Plant.id Health Assessment does not focus on a specific group of plants, although most annotations are for houseplants and ornamentals. Crop.health focuses on a broad range of diseases of selected food crops. You can read more about the differences in the crop.health FAQ section.
